You are here
Species
Florideophyceae
IUCN
NCBI
EOL Text
The Florideophyceae is a well-defined assemblage of red algae that share a number of characters including the presence of tetrasporangia, and a filamentous gonimoblast. Some species are used directly by humans for food, while cell wall polysaccharides are extracted from others for use as gels, and additives in food and cosmetic products.
| License | http://creativecommons.org/licenses/by/3.0/ |
| Rights holder/Author | D. Wilson Freshwater, Tree of Life web project |
| Source | http://tolweb.org/Florideophyceae/21781 |
While exceptions are known, members of the Florideophyceae generally share a majority of the following characteristics:
- a multicellular thallus with apical growth
- the presence of pit connections (Figure 1)
- a filamentous gonimoblast
- tetrasporangia
- cells with multiple nuclei and plastids (Figure 2)
- life histories that are some variation of a triphasic alternation of generations (Figure 3)
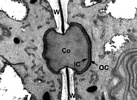
Figure 1. Electron micrograph of a Cumagloia andersonii (Nemaliales) pit plug. The pit plug occupies an aperture in the cell wall (W) and consists of a plug core (Co), flanked on either end by a thin inner cap layer (IC), and a plate-like outer cap layer (OC).
Image copyright © 2000, C.M. Pueschel.
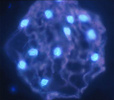
Figure 2. DAPI stained cell of Agardhiella subulata showing multiple brightly fluorescing nuclei.
Image copyright © 2000, D.F. Kapraun.
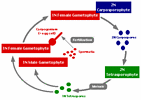
Figure 3. Diagram of triphasic life history. Haploid (1N) male gametophytes produce spermatia that are released, while haploid female gametophytes produce carpogonia (=egg cells) that are retained on the female gametophyte. After fertilization, the diploid (2N) zygote is still retained on the female gametophyte and develops into the diploid carposporophyte. The carposporophyte produces diploid carpospores that are released and develop into the diploid tetrasporophyte. The tetrasporophyte produces tetrasporangia where meiotic divisions result in haploid tetraspores. These tetraspores then develop into haploid gametophytes completing the life cycle.
Image copyright © 2000, D. W. Freshwater.
| License | http://creativecommons.org/licenses/by/3.0/ |
| Rights holder/Author | D. Wilson Freshwater, Tree of Life web project |
| Source | http://tolweb.org/Florideophyceae/21781 |
Tree based on Freshwater et al. 1994, Pueschel 1994, Ragan et al. 1994, Saunders and Bailey 1997, and unpublished analyses of S. Fredericq, D.W. Freshwater, and M.H. Hommersand.
Hypothesized relationships in the Florideophyceae are in a state of flux. Traditionally, life history characteristics, and ontogeny of the female reproductive system and carposporophyte were used to study red algal relationships. Constraints on development imposed by the filamentous construction of florideophycean taxa have lead to convergent evolution in these characters. Interpretation of these characters is therefore difficult, and in turn has prevented the establishment of solid hypotheses for higher-level relationships in the Florideophyceae.
Gabrielson and Garbary used cladistic methodology to analyze a data matrix of 35 red algal characters (Gabrielson and Garbary 1985, 1987, Garbary and Gabrielson 1990). Their resulting phylogenetic hypothesis suggested a progression from primitive (Acrochaetiales) to advanced (Ceramiales) lineages that corresponded to recognized orders.
Four of the characters in Gabrielson and Garbary's data matrix involved the ultrastructure of pit connections, which Pueschel and Cole (1982) had previously demonstrated could be used to delineate red algal orders. Pueschel (1994) subsequently developed a phylogenetic hypothesis based on these characters. In this hypothesis florideophycean taxa with two pit plug caps form a monophyletic group, and within this group, having a domed outer cap is the ancestral state (versus having plate-like outer caps).
Two overall analyses of red algae based on nucleotide sequence data from the nuclear encoded small ribosomal subunit (SSU) (Ragan et al. 1994), and the chloroplast encoded large subunit of RuBisCO (rbcL) (Freshwater et al. 1994) genes were published in 1994. While both analyses suffered from insufficient taxon sampling, they demonstrated the potential of sequence analyses for exploring the ordinal level relationships of red algae. More narrowly focused analyses of sequence data have been used to help define florideophycean orders (e.g. Fredericq et al. 1996, Saunders and Kraft 1994, 1996). Saunders and Bailey (1997) used an expanded analysis of SSU sequence data (70 taxa) to independently examine Pueschel's (1994) hypothesis of evolution based on pit connection characters, and the resulting tree provided strong support for this hypothesis. Other aspects of the Saunders and Bailey (1997) tree relating to florideophycean evolution are discussed in Saunders and Kraft (1997). More taxon replete analyses of both SSU and rbcL data are in progress and should result in a well-defined hypothesis of evolution in the Florideophyceae.
| License | http://creativecommons.org/licenses/by/3.0/ |
| Rights holder/Author | D. Wilson Freshwater, Tree of Life web project |
| Source | http://tolweb.org/Florideophyceae/21781 |
Barcode of Life Data Systems (BOLD) Stats
Specimen Records:39256
Specimens with Sequences:30909
Specimens with Barcodes:21392
Species:4387
Species With Barcodes:3847
Public Records:20243
Public Species:1885
Public BINs:2627
Genomic DNA is available from 1 specimen with morphological vouchers housed at Queensland Museum
| License | http://creativecommons.org/licenses/by-nc/3.0/ |
| Rights holder/Author | Text can be freely copied and altered, as long as original author and source are properly acknowledged. |
| Source | http://www.oglf.org/catalog/details.php?id=T01836 |